Article Text
Abstract
Background: The role of the innate immune system in the pathogenesis of asthma is unclear. Activation of innate immune receptors in response to bacterial lipopolysaccharide, viral infection and particulate matter triggers a pre-programmed inflammatory response, which involves interleukin (IL)8 and neutrophil influx. The inflammatory response in asthma is heterogeneous.
Aim: To test the hypothesis that innate immune activation may be a relevant inflammatory mechanism in neutrophilic asthma where IL8 levels are increased.
Methods: Induced sputum was obtained from non-smoking adults with asthma (n = 49), healthy controls (n = 13) and a positive reference group with bronchiectasis (n = 9). Subjects with asthma were classified into inflammatory subtypes using induced sputum cell counts. Sputum was examined for mRNA expression of the innate immune receptors toll-like receptor (TLR)2, TLR4 and CD14, and inflammatory cytokines. A separate sputum portion was dispersed and the supernatant assayed for surfactant protein A, IL8, soluble CD14 and endotoxin.
Results: Expression of innate immune receptors was increased in subjects with bronchiectasis and neutrophilic asthma compared with other asthma subtypes and controls. Increased expression of the receptors TLR2, TLR4 and CD14, as well as the pro-inflammatory cytokines IL8 and IL1β, was observed. Subjects with neutrophilic asthma had higher airway levels of endotoxin than the other groups studied.
Conclusion: There is evidence of activation of the innate immune system in asthma which results in the production of pro-inflammatory cytokines and may contribute to the pathogenesis of neutrophilic asthma.
- AHR, airway hyper-responsiveness
- APAAP, alkaline phosphatase anti-alkaline phosphatase
- DTT, dithiothreitol
- FEV1, forced expiratory volume in 1 s
- LAL, Limulus Amebocyte Lysate
- LPS, lipopolysaccharide
- PAMP, pathogen-associated molecular pattern
- PCR, polymerase chain reaction
- SP-A, surfactant protein A
- TLR, toll-like receptor
- TNF, tumour necrosis factor
Statistics from Altmetric.com
- AHR, airway hyper-responsiveness
- APAAP, alkaline phosphatase anti-alkaline phosphatase
- DTT, dithiothreitol
- FEV1, forced expiratory volume in 1 s
- LAL, Limulus Amebocyte Lysate
- LPS, lipopolysaccharide
- PAMP, pathogen-associated molecular pattern
- PCR, polymerase chain reaction
- SP-A, surfactant protein A
- TLR, toll-like receptor
- TNF, tumour necrosis factor
The role of the innate immune system in the pathogenesis of asthma is unclear, but may be relevant to the heterogeneous inflammatory response that occurs in asthma.1–,5 Acquired immune responses in asthma are well characterised and involve allergen-induced T helper type 2 lymphocyte activation and consequent eosinophilic airway inflammation. Activated eosinophils release potent cytotoxic granules such as major basic protein and eosinophil cationic protein which induce airway hyper-responsiveness (AHR) and symptoms.6,7 Recently, non-eosinophilic inflammatory subtypes of asthma have been identified3,4,8,9,10,11,12,13,14,15,16 where symptoms and AHR persist in the absence of increased sputum eosinophils. The mechanisms of non-eosinophilic asthma and, more particularly, neutrophilic asthma are not well characterised; however, a potential role for neutrophils and interleukin (IL)8 has been reported.3,16 IL8-mediated neutrophil influx often occurs with nuclear factor κB activation, and represents a “pre-programmed” response that has been conserved throughout evolution,5 and is typically seen with activation of the innate immune system.17 This suggests that neutrophilic asthma may involve activation of the innate immune system.
The innate immune system is rapidly activated by pathogen-associated molecular patterns (PAMPs). PAMPs such as lipopolysaccharide (LPS) are highly conserved structures common to many microorganisms. They are recognised by pattern-recognition receptors such as the toll-like receptors (TLRs), CD14 and collectins, which include pulmonary surfactant proteins.17 TLR activation triggers a signalling cascade leading to the activation and nuclear translocation of nuclear factor κB, resulting in a pro-inflammatory cytokine response including tumour necrosis factor α (TNFα), IL8 and IL1β. 1,18
This study questioned whether activation of the innate immune system was a feature of neutrophilic asthma and tested the hypothesis that subjects with asthma and a neutrophilic inflammatory subtype would have activation of the innate immune response characterised by increased expression of innate pattern-recognition receptors TLR2, TLR4, surfactant protein A (SP-A) and CD14, and a corresponding cytokine response. In addition, we assessed whether levels of sputum LPS and bacteria were associated with asthma subtype.
METHODS
Subjects and design
A cross-sectional study design was used. Non-smoking subjects with asthma (n = 49, American Thoracic Society criteria)19 had a clinical diagnosis of symptomatic asthma and AHR to hypertonic saline. Controls (n = 13) without respiratory disease had a forced expiratory volume in 1 s (FEV1) >80% of predicted20 and normal airway responsiveness. Subjects with bronchiectasis (confirmed by high-resolution CT, n = 9) were recruited as a positive reference group. All subjects were stable (no lower respiratory tract infection or exacerbation of respiratory disease in the previous 4 weeks) at the time of assessment. Subjects were recruited from the Respiratory Ambulatory Care Service, John Hunter Hospital, New Lambton, New South Wales, Australia, and by advertisement, and gave written informed consent. They underwent clinical assessment, spirometry, combined hypertonic saline challenge and sputum induction.21 Those with neutrophilic asthma underwent high-resolution CT scanning to exclude the presence of coexisting bronchiectasis. The Hunter Area Health Service and the University of Newcastle research ethics committees approved this study.
Sputum induction
Spirometry (KoKo PD Instrumentation, Louisville, Colorado, USA) and combined bronchial provocation testing and sputum induction with hypertonic saline (4.5%) were performed as described previously.21 Sputum was induced using normal (0.9%) saline in 12 (23%) subjects with asthma and two (22%) subjects with bronchiectasis where the post-bronchodilator FEV1 was <1.5 l. A fixed sputum induction time of 15 min was used for all subjects.
Sputum analysis
Selected sputum (100 μl) was transferred to RNA extraction buffer (Qiagen, Hilden, Germany) and stored at −80°C. RNA was prepared as described below (see also Simpson et al16). RNA purity and titre were determined by spectrophotometry (Cary 50; Varian, Palo Alto, California, USA) and 100 ng RNA was applied to subsequent reverse transcription-polymerase chain reactions (PCR).
RNA preparation
RNA was prepared using the RNeasy Mini Kit (Qiagen). Random primers were combined with sample RNA and a 20 μl reverse transcription mix containing 0.5 mM deoxyribonucleotide triphosphates, 5 mM dithiothreitol (DTT), 50 mM TRIS-hydrochloride, pH 8.3, 75 mM potassium chloride and 2 U recombinant ribonuclease inhibitor. After incubation, the mix was supplemented with 100 U Superscript II RNase and incubated at 42°C for 50 min. After enzyme inactivation, the cDNA was stored at −20°C. All reverse transcription-PCR reagents other than deoxyribonucleotide triphosphates (Promega Corporation, Madison, Wisconsin, USA) were purchased from Invitrogen (Carlsbad, California, USA).
RNA extraction and reverse transcription
Primers for TLR222 were custom made (Qiagen) and the probe was designed using Primer V.3 software (http://www.premierbiosoft.com/netprimer/netprimer.html). All other primers and probes were purchased in kit form (Applied Biosystems, Foster City, California, USA).
Semi-quantitative real-time PCR
Multiplex PCR of each target and the endogenous reference (18-S ribosomal RNA) was performed containing 2 μl of the sample cDNA, 300 nM each of target primers, 100 nM of probe and a commercial polymerase solution (Universal PCR Master Mix, Applied Biosystems). Each sample was subject to amplification with the following parameters: 2 min at 50°C, 10 min at 95°C and 40 cycles of 95°C for 15 s followed by 1 min at 60°C (ABI GeneAmp 7700 cycler, Perkin-Elmer, Foster City, California, USA). The amount of target was calculated relative to a positive calibrator expressing the target of interest and normalised to the endogenous control (18-S).23
Specific target positive calibrators
Peripheral blood mononuclear cells and granulocytes were isolated using Percoll separation (Amersham, Castle Hill, New South Wales, Australia) and cultured in the presence of LPS (100 ng/ml, Sigma, St Louis, Missouri, USA) or phytohaemagglutinin (5 μg/ml, Sigma) for 6 h at 37°C, 5% CO2. Stimulated cells were stored in RNA extraction buffer at –80°C for RNA extraction.
An aliquot of sputum was used for bacteriological culture. Chocolate and blood agar plates were inoculated with induced sputum (10 μl) and transported with prepared sputum smears for Gram staining and identification to the microbiology department of the Hunter Area Pathology Service for culture and reporting. The remaining selected sputum was dispersed using DTT as described previously.21 The suspension was filtered and a total cell count of leucocytes and viability performed. After centrifugation, the supernatant was aspirated and stored at −80°C. Cytospins were prepared, stained (May–Grunwald Geimsa) and a differential cell count obtained from 400 non-squamous cells. TLR protein expression was assessed in sputum cytospins (alkaline phosphatase anti-alkaline phosphatase (APAAP) technique) with anti-human TLR2 (Clone TLR2.1) and TLR4 (Clone HTA125) antibodies (Serotec, Oxford, UK) as described previously.24 Cytospins were fixed using paraformaldehyde, l-lysine, periodate and allowed to air dry before being immersed in sucrose solution for storage.25 Slides were stored at −20°C until immunocytochemical analysis was undertaken. They were then thawed and washed in TRIS buffered saline (H+S), and blocked using 20% normal rabbit serum (Dako, Botany, NSW, Australia) for 20 min. Primary antibody was applied and the slides were incubated at room temperature for 30 min. They were then washed before application of link rabbit anti-mouse immunoglobulins (Dako). Mouse monoclonal APAAP (Dako) was applied after removal of excess link antibody by washing. Excess APAAP was removed by washing before application of liquid permanent red substrate (Dako). A substrate control was included, which had TRIS buffered saline (H+S) applied at each step until the substrate was added. Slides were observed for colour development and substrate action was stopped by a distilled water wash, when the positive control had developed a strong red colour. A haematoxylin counterstain was applied and the slides were air dried before being coverslipped using DPX (Sigma).
Positive control slides
Peripheral blood granulocytes were isolated using Percoll centrifugation (Amersham, Uppsala, Sweden) and stimulated with 100 ng/ml LPS (TLR4) or granulocyte macrophage colony-stimulating factor (TLR2) for 2 h. Cytospins prepared from the cell suspension were fixed in paraformaldehyde, l-lysine, periodate and allowed to air dry before being immersed in sucrose solution for storage.25 Slides were stored at −20°C for subsequent immunocytochemical analysis.
Asthma subtype classification
The upper limit of normal for sputum eosinophil and neutrophil counts was taken as the 95th centile of a healthy control population studied.16 Subjects with a sputum neutrophil count ⩾61% were classified as having neutrophilic asthma, those with sputum eosinophils ⩾1% alone were classified as having eosinophilic asthma and those with sputum neutrophils <61% and sputum eosinophils <1% were classified as having paucigranulocytic asthma. On the basis of prior work, subjects with both increased neutrophils and eosinophils were classified as having neutrophilic asthma.16
Sputum fluid-phase mediator assessment
Determination of IL8 was by ELISA (R&D Systems, Minneapolis, Minnesota, USA). Active neutrophil elastase was measured using a chromogenic substrate specific for human neutrophil elastase, n-methoxysuccinyl-l-alanyl-l-alanyl-prolyl-l-valyl-p-nitroanilide (Sigma), as described previously.26 Both IL8 and neutrophil elastase have been previously validated for assessment in induced sputum.27 SP-A concentration was determined by adapting a previously described method.28 SP-A antigen and antibody were a gift from Dr Ian Doyle, Flinders University, South Australia, Australia. The mean recovery of SP-A (110 ng/ml) added to sputum supernatant processed using DTT dispersion was 84% (range 70–99%). No effect of DTT on the standard curve was observed. Good agreement was found in the SP-A concentrations measured in sputum supernatants diluted 1 in 12 or 1 in 24 with reagent diluent (phosphate-buffered saline bovine serum albumin, n = 8), with all samples within the Bland–Altman limits of agreement (mean bias (2SD)). The coefficient of variance was 40.6%. Supernatant measurements were performed with coded samples by a technician blinded to the patient’s inflammatory group.
Airway endotoxin
Endotoxin in sputum supernatant was assayed with a quantitative kinetic chromogenic Limulus Amoebocyte Lysate (LAL) method (Kinetic QCL number 50-650 U; Bio Whittaker; LAL Lot number 3L2360; CSE Lot number 2L4900 and Lysate Lot number 3L085D) at 37°C.29 Inhibition or enhancement of the LAL assay was not detectable at sample dilutions of 50 times or higher, tested as described previously.30 Owing to limited supernatant samples, endotoxin levels were assessed in samples with neutrophilic asthma (n = 6), eosinophilic asthma (n = 24), paucigranulocytic asthma (n = 10), controls (n = 6) and all bronchiectasis samples.
Data analysis
Data were analysed using Stata V.7, with results reported as median and interquartile range unless otherwise indicated. Analysis was performed using the two-sample Wilcoxon rank sum test or Kruskal–Wallis test for more than two groups with Bonferroni correction. Fisher’s exact test was used to analyse categorical data. Associations between data were determined using Spearman’s rank correlation. Results were reported as significant when p<0.05.
RESULTS
Clinical features
Of the 49 subjects with asthma, seven (14%) had neutrophilic asthma, 26 (53%) had eosinophilic asthma and 16 (33%) had paucigranulocytic asthma. The subjects with asthma had similar clinical characteristics across the three inflammatory subtypes (table 1⇓). The severity of air flow obstruction was similar in subjects with asthma and bronchiectasis, and AHR was present in subjects with asthma, but not in controls or in those with bronchiectasis (table 1⇓). Three subjects in the neutrophilic asthma group had a mixed granulocytic pattern of inflammation with increased neutrophils and eosinophils.
Clinical characteristics of subjects with neutrophilic, eosinophilic and paucigranulocytic asthma, bronchiectasis and controls
Although not statistically significant, subjects with neutrophilic asthma tended to have a lower FEV1 % predicted and FEV1/FVC than subjects with eosinophilic asthma and paucigranulocytic asthma (table 1⇑).
Inflammatory cells
Subjects with neutrophilic asthma had an increased total cell count and an increased proportion and number of neutrophils compared with subjects with eosinophilic asthma, paucigranulocytic asthma and controls (table 2⇓). Absolute macrophage counts were similar between groups, indicating that the reduced proportion of macrophages occurred because of the high neutrophil proportion and the expression of the results as a percentage. No correlation with sputum neutrophils and age was observed. Patients with eosinophilic asthma had an increased proportion and number of eosinophils and increased neutrophil proportion compared with those with paucigranulocytic asthma. Subjects with paucigranulocytic asthma showed no differences in inflammatory cell counts from controls.
Inflammatory cell counts for subjects with neutrophilic, eosinophilic and paucigranulocytic asthma, bronchiectasis and controls
Innate immune receptor expression
Increased mRNA expression of TLR2, TLR4 and CD14 was observed in those with neutrophilic asthma compared with those with eosinophilic and paucigranulocytic asthma. SP-A levels were also significantly higher in subjects with neutrophilic and eosinophilic asthma than in controls (fig 1⇓). Expression of TLR2 and CD14 mRNA was significantly higher in those with neutrophilic asthma than in controls. Although mRNA expression of TLR2, TLR4 and CD14 was higher in subjects with neutrophilic asthma, these subjects were not statistically different from the positive reference subjects with bronchiectasis, who had increased TLR2 mRNA and SP-A protein in induced sputum compared with controls (p<0.05, fig 1⇓). No differences in soluble CD14 levels were found between any subject groups studied. TLR immunocytochemistry indicated the presence of TLR2 and TLR4 on sputum macrophages and neutrophils.
Sputum innate pattern recognition receptors in subjects with neutrophilic asthma, eosinophilic asthma, paucigranulocytic asthma, bronchiectasis and in controls. Horizontal bars represent median values. p<0.0025 (Kruskal–Wallis) versus *eosinophilic asthma, †paucigranulocytic asthma and ‡controls. (A) Toll-like receptor (TLR)2 mRNA expression; (B) TLR4 mRNA expression; (C) surfactant protein A (SP-A) protein levels; (D) CD14 mRNA expression.
Subjects with eosinophilic asthma had increased levels of SP-A compared with controls (fig 1⇑), but similar levels of all other innate immune receptors and inflammatory markers (figs 1⇑ and 2⇓). Subjects with paucigranulocytic asthma were similar to controls for all of the innate immune activation and inflammation markers assessed.
Sputum proinflammatory cytokines in subjects with neutrophilic asthma, eosinophilic asthma, paucigranulocytic asthma, bronchiectasis and in controls. Horizontal bars represent median values. p<0.0025 versus *eosinophilic asthma, †paucigranulocytic asthma and ‡controls, respectively. (A) Interleukin (IL)8 mRNA expression; (B) IL8 protein levels; (C) tumour necrosis factor (TNF)α mRNA expression; (D) IL1β mRNA expression.
Innate immune cytokines
Signature cytokines of innate immune activation assessed included IL8, IL1β and TNFα. Subjects with neutrophilic asthma had increased mRNA expression of IL8, TNFα and IL1β, as well as increased IL8 protein compared with those with paucigranulocytic asthma. Although IL1β and TNFα mRNA levels were higher in neutrophilic asthma, only IL8 mRNA levels were significantly increased in comparison with eosinophilic asthma. Fluid phase protein levels and mRNA expression of IL8 were significantly increased in those with neutrophilic asthma and bronchiectasis compared with controls (fig 2⇑). IL1β and TNFα mRNA expression tended to be higher in those with neutrophilic asthma and bronchiectasis than in controls; however, these levels did not reach statistical significance. IL1β and TNFα protein levels could not be assessed in the sputum supernatant owing to the effect of DTT on measurement of these cytokines.
Neutrophil elastase was detected in 67% of sputum samples from subjects with neutrophilic asthma compared with 4% from those with eosinophilic asthma (p<0.002, fig 3⇓). No samples from subjects with paucigranulocytic asthma or controls had detectable neutrophil elastase. A similar proportion of subjects with bronchiectasis had neutrophil elastase in their sputum supernatant (67%).
Neutrophil elastase (NE) detected in sputum supernatant from subjects with asthma and bronchiectasis and from controls.
Innate immune agonists
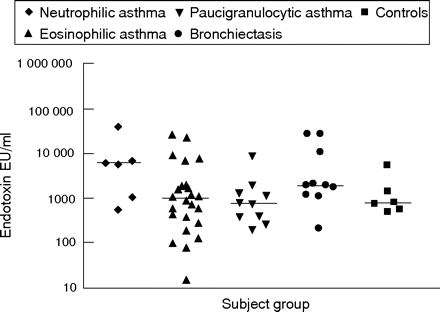
Endotoxin and bacterial colonisation were examined as potential activators of the innate immune response. Airway endotoxin levels were high in neutrophilic asthma (median 5797 endotoxin units (EU)/ml, fig 4⇓) and tended to be higher than in subjects with eosinophilic asthma (median 955 EU/ml) and paucigranulocytic asthma (median 729 EU/ml); however, these results did not reach statistical significance. Similarly, levels were lower in subjects with bronchiectasis (median 1807 EU/ml) and in controls (median 738 EU/ml).
Airway endotoxin levels in sputum supernatant from subjects with neutrophilic, eosinophilic and paucigranulocytic asthma, bronchiectasis and from controls. Horizontal bars represent median levels. EU, endotoxin units.
A significant negative correlation was observed between airway endotoxin levels and airway obstruction (FEV1/FVC%, r = −0.38, p = 0.002). Also, sputum neutrophil proportion (r = 0.33, p = 0.035) and IL8 protein levels (r = 0.33, p = 0.036) were positively associated with airway endotoxin in subjects with asthma (table 3B⇓).
Correlation matrix for innate immune activation receptors (A) and inflammatory response markers (B) in patients with asthma
Subjects with neutrophilic asthma had a greater prevalence of airway bacterial colonisation (43% with a positive bacteria culture) than those with other asthma subtypes (8% and 0% eosinophilic and paucigranulocytic, respectively, tables 1⇑ and 4⇓). Five of the nine subjects with bronchiectasis studied had evidence of chronic colonisation by common Gram negative respiratory pathogens (Haemophilus influenzae and Pseudomonas aeruginosa). No subject returned positive PCR results for human rhinovirus or respiratory syncytial virus (data not shown).
Bacteria identified in subjects with asthma
Associations
TLR2 mRNA levels were associated with TLR4 and CD14 mRNA (table 3A⇑) and also correlated with IL8 mRNA and protein, TNFα, IL1β mRNA and the neutrophil proportion. TLR4 mRNA levels were strongly associated with CD14 mRNA and TNFα mRNA (table 3⇑). Weaker significant correlations were observed with IL8 mRNA and the neutrophil proportion. SP-A levels were significantly associated with total cell count (table 3⇑) and weakly associated with IL8 protein levels (table 3⇑). IL8 mRNA levels were significantly associated with IL8 protein levels, IL1β and TNFα mRNA levels (table 3⇑). No correlation was found between CD14 mRNA levels and soluble CD14 protein levels (p>0.05). Endotoxin levels in sputum supernatant were positively associated with the sputum neutrophil proportion and IL8 protein levels (table 3B⇑).
DISCUSSION
This is the first clinical study to examine innate immune responses in the airways of subjects with asthma and bronchiectasis. Neutrophilic asthma was associated with an upregulation of the innate immune response. In particular, we found that neutrophilic asthma was characterised by increased expression of several key innate immune receptors: TLR2, TLR4, CD14 and SP-A, as well as pro-inflammatory cytokines IL8 and IL1β. We also found high levels of airway endotoxin in subjects with neutrophilic asthma. Innate immune activation may therefore be a key mechanism in the development of neutrophilic asthma.
Subjects with bronchiectasis were used in this study to provide a positive reference group, and this represents the first report of innate immune markers in bronchiectasis. These subjects were expected to have innate immune activation owing to their chronic bacterial infection and chronic airway inflammation with neutrophils. Persistent bacterial colonisation of the airways in bronchiectasis is caused by impaired mucociliary transport and mucus clearance, and initiates a vicious cycle of inflammation characterised by activated neutrophils and neutrophil proteases.31 On persistent exposure to PAMPs, we hypothesised and confirmed that innate receptors would be stimulated and activated to induce the production of IL8 and an influx of neutrophils. Subjects with bronchiectasis had increased sputum mRNA expression of TLR2 and supernatant SP-A compared with controls. No difference was observed in TLR4 mRNA expression, suggesting that there may be differential expression of TLRs in bronchiectasis.
Innate immune activation seems to be an important pro-inflammatory mechanism in neutrophilic asthma. While the pathway leading to eosinophilic asthma is well characterised, the mechanisms of neutrophilic inflammation in asthma are poorly understood. Although neutrophils were thought to be primarily involved in severe asthma,32,33 increased neutrophil levels have also recently been reported in stable asthma.16,34,35 The increased neutrophil levels in those studies were not caused by respiratory tract infections as subjects with a reported lower respiratory tract infection in the preceding month were excluded. In the study by Green et al,35 subjects with neutrophilic asthma were older, had a later onset of disease and were less atopic than subjects with normal levels of neutrophils. Our study was too small to assess these differences. However, it is important to note that neutrophilic asthma has been described in people with mild, moderate and severe asthma, and it is not merely a feature of severe asthma with fixed air flow obstruction.6,13
Sputum TNFα mRNA levels were also highest in subjects with neutrophilic asthma and bronchiectasis, and TNFα mRNA was significantly correlated with both TLR4 and TLR2 mRNA expressions. Recently, TNFα has been implicated in the upregulation of TLR2 expression in epithelial cells and, therefore, may be an important cytokine in the perpetuation of innate immune activation in the airways.32 Further, a recent randomised controlled trial of a soluble TNFα receptor in people with severe refractory asthma has shown improvements in a number of asthma outcomes including asthma control score and AHR, highlighting the potentially important pathogenic role of TNFα in difficult asthma.33
The inflammatory cell counts in neutrophilic asthma were similar to bronchiectasis and, in addition, both groups had an increased frequency of chronic bacterial colonisation of the airways when compared with the other asthma subtypes. This indicates that neutrophilic asthma and bronchiectasis have a similar pattern of airway inflammation, with evidence of innate immune activation. Sputum endotoxin levels were high in subjects with neutrophilic asthma, which suggests that endotoxin may be the source of PAMPs driving the innate response in this group. The levels were 6–8-fold higher than those observed in the other asthma subtypes studied. In contrast, to subjects with bronchiectasis, there was increased mRNA expression of both TLR2 and TLR4 in subjects with neutrophilic asthma compared with both eosinophilic and paucigranulocytic asthma. Although TLR4 has been identified as the primary receptor for bacterial LPS, recent studies have shown that TLR2 is also capable of responding to LPS.36 This may explain why both TLR2 and TLR4 were upregulated. Alternatively, other PAMPs or cytokines known to upregulate TLR2 expression may have a role.
LPS signalling is complex and involves a number of accessory proteins including LPS binding protein and CD14. Subjects with neutrophilic asthma had increased mRNA expression of CD14 and TLR4 compared with controls, subjects with eosinophilic and paucigranulocytic asthma and those with bronchiectasis. The reason for this increase in neutrophilic asthma (which was not observed in bronchiectasis) is unclear, but may be due to the higher levels of endotoxin measured in the sputum supernatant, which may be upregulating the expression of LPS signalling proteins. There was also a strong positive correlation between TLR4 mRNA and CD14 mRNA in subjects with asthma, suggesting that the expression of these proteins is co-regulated.
The roles of gene polymorphisms in determining innate immune responses are conflicting. The TLR4 polymorphism Asp299Gly has been associated with hyporesponsiveness to airway challenge with LPS37 and increased risk of Gram-negative infection.38 In another study, similar rates of chronic infection were reported between the subjects with a TLR4 polymorphism and wild-type TLR4, indicating a role for this polymorphism in the risk of acute infection but not in chronic infection.39 The same TLR4 polymorphism has been associated with asthma in Swedish children,40 but no association was found between TLR4 polymorphisms and a diagnosis of asthma in a large adult study.41 A polymorphism in the CD14 promoter has been associated with increased soluble CD14 expression and lower IgE levels in serum,42 whereas polymorphisms of the LPS-binding protein gene have been associated with increased risk of sepsis in association with male gender.43 Regardless of these conflicting data, it is plausible that a polymorphism in innate immune receptors and associated proteins (CD14 and LPS-binding protein) may explain the variation in immune and inflammatory responses reported.
SP-A functions primarily as an opsonin, identifying targets for phagocytosis and binding of pathogens in vitro. In SP-A null mice, exposure to pathogens, including viruses, results in increased neutrophilia, epithelial injury and persistence of infection compared with control mice, indicating a protective role for surfactant proteins in lung defence.44 In this study, levels of SP-A were increased in subjects with eosinophilic and neutrophilic asthma compared with controls, indicating host-defence activity in these subjects. SP-A did not seem to be a specific marker of innate immune activation as levels were similar between the asthma subtypes. Although levels of SP-A are increased, it is unknown whether the SP-A is functional and intact. Neutrophil proteases present in airway fluids degrade SP-A and reduce its antimicrobial and anti-inflammatory functions. SP-A levels are also increased in bronchial lavage fluid from subjects with asthma.45 Further investigation is required to determine whether the integrity of SP-A from the airways of subjects with asthma is maintained.
In summary, we conclude that persistent activation of the innate immune system in stable asthma results in the production of pro-inflammatory cytokines which may contribute to the pathogenesis of neutrophilic asthma. This is an important observation that identifies a specific mechanism operating in the neutrophilic subtypes of non-eosinophilic asthma. This adds to our understanding of the heterogeneity of airway inflammation in asthma. The mechanisms of this persistent activation are unclear but may be related to endotoxin exposure or chronic bacterial colonisation of the lower airways.
Acknowledgments
We thank Ms Naomi Timmins, Ms Rebecca Oldham, Ms Debbie Pepperal, Ms Suzanne Meldrum and Ms Noreen Bell for their technical contributions.
REFERENCES
Footnotes
Published Online First 14 July 2006
Funding: This study was funded by NHMRC and Hunter Medical Research Institute, New South Wales, Australia.
Competing interests: None.